- We are interested in understanding the 3D genome organization, starting from base pair level to the whole genome in single cells.
- We develop and apply super-resolution imaging technologies to understand the global organization of cellular structures with a main focus on the functional genome.
- Using single-molecule imaging technologies, we focus to understand the enzymes that shape the genome.
Measuring DNA energetics for understanding genome maintenance
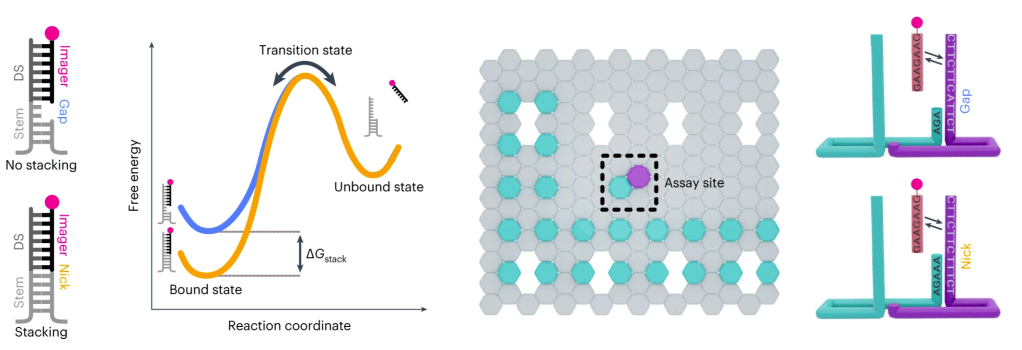
Single-molecule assay for measuring individual dinucleotide base-stacking energetics and representative free-energy diagram showing base-stacking energy. A graphic of the origami layout with the L-shaped cyan-colour extensions identifies the locations of origami structures, and the assay site with magenta-colour extension to study base-stacking interactions. Right: detailed view of the assay site. Ref: Banerjee et al., Nature Nanotechnology, 2023.
The DNA double helix structure is fundamentally stabilized by two primary mechanisms: base-pairing and base-stacking interactions. These interactions are essential not only for maintaining the integrity of the DNA structure but also for ensuring proper biological function. We are developing advanced single-molecule imaging assays aimed at gaining a comprehensive understanding of dinucleotide base-stacking energetics, which play a crucial role in the overall stabilization of DNA. By combining the sophisticated technique of DNA-PAINT imaging with rationally designed DNA nanostructures, we can effectively measure the free energy associated with individual dinucleotide stacking interactions. Such precise measurements are vital, as they will significantly contribute to our ability to design functional DNA nanostructures and aptamers with enhanced capabilities. Ultimately, these findings will facilitate a greater understanding of the intricate role that local DNA energetics play during critical processes like DNA repair, opening new avenues for research and potential therapeutic applications in genetic stability and cellular health.
Related publications:
1) Banerjee A., et al., Single-molecule analysis of DNA base-stacking energetics using patterned DNA nanostructures, Nature Nanotechnology, Aug 2023
2) Banerjee A., Anand M. et al, labeling approaches for DNA-PAINT super-resolution imaging, Nanoscale, Mar 2023
DNA compaction by nucleoid-associated proteins at single-molecule level
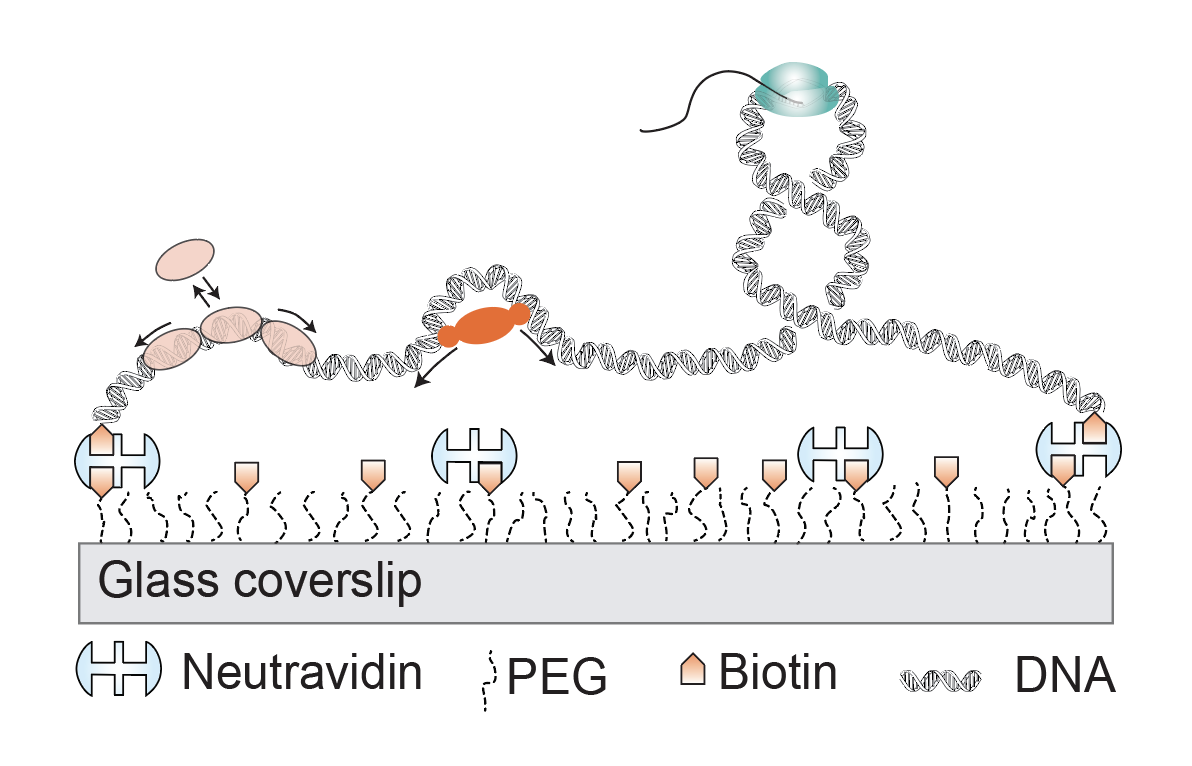
In this project, we explore DNA-protein interactions using single-molecule imaging techniques. Our main focus is to understand the enzymes that bind DNA to protect, copy, and/or modulate the conformation to organize the genome. For instance, we study mechanism of functioning of Nucleoid-Associated Proteins (NAPs) and their influence on transcription. We showed that Dps, a NAP that is expressed in E. coli under starvation or stress, compacts DNA by bridging of neighboring strands. Another NAP from Mycobacterium, called Lsr2, that plays a key role in regulating horizontally acquired genes is appeared to form phase-separated condensates that can scavenge large number of genes specifically.

A Nucleoid-Associated Protein called Lsr2 from Mycobacterium Tuberculosis compacts DNA by sequence-dependent co-condensation thereby simultaneously binds to large number of specific genes for regulating their expression.
Related publications:
1) Gaur P. et al., Sequence-dependent co-condensation of Lsr2 with DNA elucidates the mechanism of genome compaction in Mycobacterium tuberculosis, Biorxiv, 2025
2) Shahu S., et al, Bridging DNA contacts allow Dps from E. coli to condense DNA, Nucleic Acids Research, 2024
Understanding nanoscale genome organization using multiplexed DNA-PAINT

Nanoscale genome organization using Multiplexed DNA-PAINT. Antibodies against each of the epigenetic modifications are conjugated with unique DNA strands via secondary nanobody. After antibody labeling of all targets in cells, each of the target is imaged using DNA-PAINT in sequential fashion. Multiplexed DNA-PAINT revealing that transcription inhibition leads to a dramatic reorganization of genome. Ref: Banerjee et al., Biorxiv, 2025.
DNA-Points Accumulation in Nanoscale Topology (DNA-PAINT) technique is one of the single-molecule localization microscopy (SMLM) based techniques. In DNA-PAINT, the targets of interest are tagged with short, unique (<9 nt) DNA barcode called docking strands. The targets are then imaged sequentially using fluorophore-conjugated complementary imager strands. The imager strand binds the docking strand transiently bind to create the necessary blinking events to create super-resolution images. Each of the cellular targets are typically targeted with DNA docking strand conjugated antibodies and imaged with corresponding imager strands. DNA Fluorescence In situ Hybridization (DNA-FISH) in combination with DNA-PAINT allow us to study the genome architecture and its role in gene regulation. We are applying DNA-PAINT technique to image the nuclear architecture in mammalian cells, cell division, and many other fascinating biological phenomenon.
Related publications:
1) Banerjee A., Anand M. et al., High-Speed Multiplexed DNA-PAINT Imaging of Nuclear Organization Using an Expanded Sequence Repertoire, Biorxiv, 2025